Background
Mechanism Syntax
The official example is saved on the GitHub:
---
species_list:
CL: Cl
O1D: O
'NO': N + O
ALK4: 4*C
...
reaction_list:
IRR_1: O3 + NO ->[k] 1.000*NO2
IRR_2: NO2 ->[j] 1.000*NO + 1.000*O
IRR_3: O2 + O ->[k] O3
...
species_group_list:
- NOx = NO + NO2
- NOz = HNO3 + RNO3
- PANS = PAN + HPAN + GPAN
- NOy = NOx + 2*N2O5 + NO3 + HNO4 + NOz + PANS
...
I don’t care about the species_list and set it to ignore:
species_list:
'CL': IGNORE
The most important part is reaction_list.
Reactions are combinations of species, stoichiometry, reaction role, and reaction type. Each reaction is identified by a reaction label, which can be any YAML recognizable string that starts with a letter and contains only letters and numbers. The reaction definition starts with a reactants list containing one or more reactants with optional stoichiometry. The reactants list is followed by an arrow (i.e.
->) with the reaction type enclosed in braces (i.e.[k]or[j]) where j specifies photolysis and k specifies thermal. The reaction arrow and type are followed by an optional products list that has the same syntax as the reactants list.
The reaction label should be the variable names saved in the IRR* files. For WRF-Chem, the name list is saved in Registry/registry.irr_diag:
state real - ikjf irr_diag_mozcart - - - - "Integrated Reaction Rate" ""
state real M_HV_IRR ikjf irr_diag_mozcart 1 - rh9 "M_HV_IRR" "M+HV Integrated Reaction Rate" "ppmv"
state real O3_HV_IRR ikjf irr_diag_mozcart 1 - rh9 "O3_HV_IRR" "O3+HV Integrated Reaction Rate" "ppmv"
state real O3_HV_a_IRR ikjf irr_diag_mozcart 1 - rh9 "O3_HV_a_IRR" "O3+HV_a Integrated Reaction Rate" "ppmv"
state real N2O_HV_IRR ikjf irr_diag_mozcart 1 - rh9 "N2O_HV_IRR" "N2O+HV Integrated Reaction Rate" "ppmv"
state real NO2_HV_IRR ikjf irr_diag_mozcart 1 - rh9 "NO2_HV_IRR" "NO2+HV Integrated Reaction Rate" "ppmv"
state real N2O5_HV_IRR ikjf irr_diag_mozcart 1 - rh9 "N2O5_HV_IRR" "N2O5+HV Integrated Reaction Rate" "ppmv"
...........
The rest part is the reaction equations in chem/KPP/mechanisms/.
I will focus on mozcart mechanism which corresponds to the eqn file called chem/KPP/mechanisms/mozcart/mozcart.eqn:
#EQUATIONS {MOZART, check troee, usr5, usr17, rc_n2o5 }
// photorates
{001:J01} M{=O2}+hv=O+O : .20946_dp*j(Pj_o2) ;
{002:J02} O3+hv=O1D_CB4{+O2} : j(Pj_o31d) ;
{003:J03} O3+hv=O{+O2} : j(Pj_o33p) ;
{004:J04} N2O+hv=O1D_CB4{+N2} : j(Pj_n2o) ;
{005:J05} NO2+hv=O+NO : j(Pj_no2) ;
{006:J06} N2O5+hv=NO2+NO3 : j(Pj_n2o5) ;
{007:J07} HNO3+hv=OH+NO2 : j(Pj_hno3) ;
{009:J09} NO3+hv=.89 NO2+ .11 NO + .89 O3: j(Pj_no3o) ;
................
Set up the reaction_list
Steps
- Get the reaction label from the
registry.irr_diagdirectly, as they’re in the same order. - Generate the equation from the texts between the first
}and the second:from themozcart.eqn:- Drop spaces which are not in the equation part
- Replacing the spaces with
* - Neglect
{***}in the texts - Replace
+hv=with->[j] - Replace
=with->[k]
Script
import pandas as pd
eqn = pd.read_csv('./mozcart.eqn', header=None, sep=':', comment='/', skiprows=1)
registry = pd.read_csv('./registry.irr_diag', header=None, delim_whitespace=True, skiprows=1)
eqn_clean = eqn[1].str.replace(r"\{.*?\}","").str.replace(r".*?\}","")\
.str.strip().str.replace(" ", "*")\
.str.replace("\+hv=", "->[j]")\
.str.replace("=", "->[k]")
registry_mozcart = registry[registry[4] == 'irr_diag_mozcart']
eqn[0] = registry_mozcart[8]
eqn[1] = eqn_clean
eqn = eqn.drop(columns=[2])
tsv = eqn.to_csv(sep=':', header=False, index=False)
psv = tsv.replace(':', ': ')
with open('./mozart_wrfchem.yaml', 'w') as outfile:
outfile.write(psv)
Result
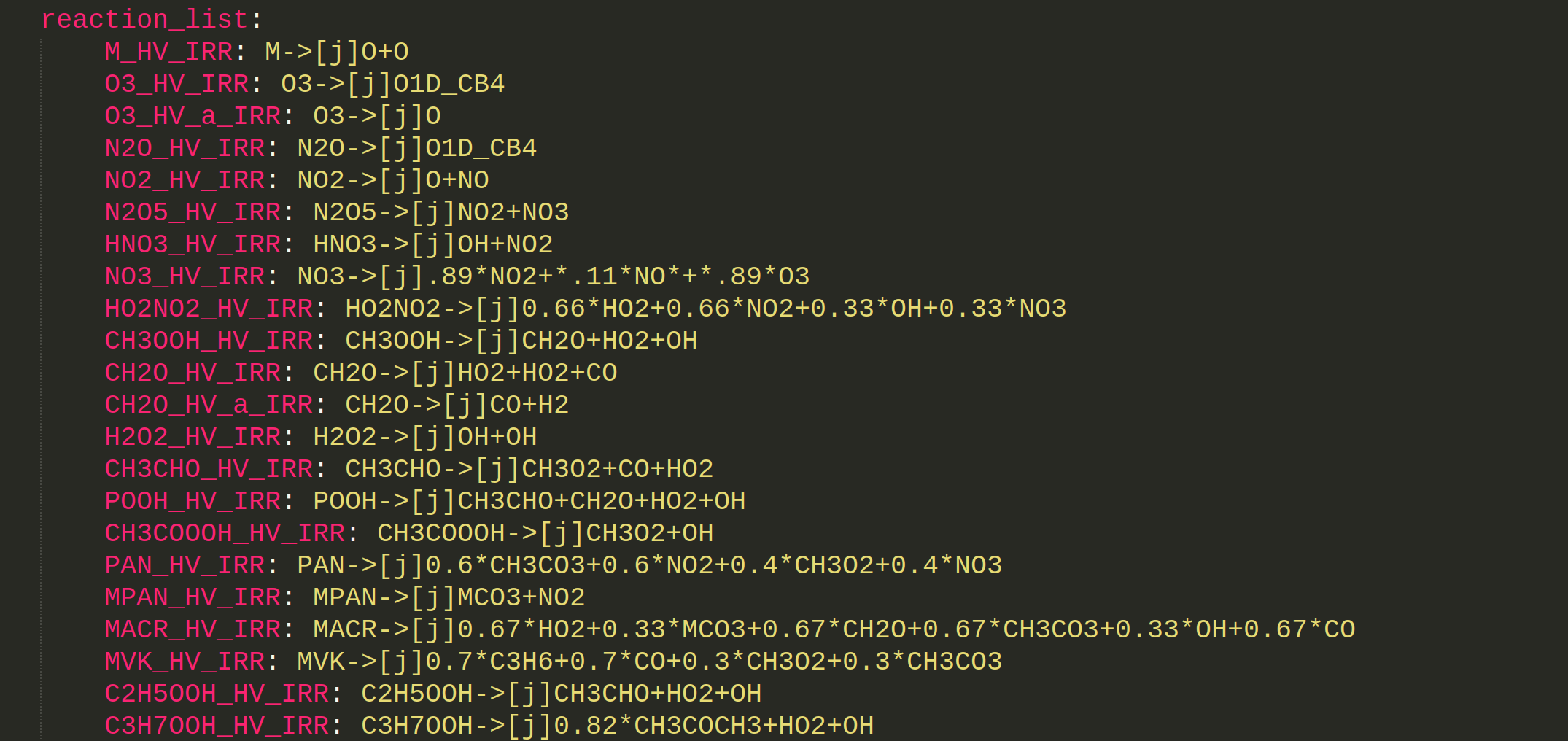
M_HV_IRR: M->[j]O+O
O3_HV_IRR: O3->[j]O1D_CB4
O3_HV_a_IRR: O3->[j]O
N2O_HV_IRR: N2O->[j]O1D_CB4
NO2_HV_IRR: NO2->[j]O+NO
N2O5_HV_IRR: N2O5->[j]NO2+NO3
HNO3_HV_IRR: HNO3->[j]OH+NO2
NO3_HV_IRR: NO3->[j].89*NO2+*.11*NO*+*.89*O3
HO2NO2_HV_IRR: HO2NO2->[j]0.66*HO2+0.66*NO2+0.33*OH+0.33*NO3
CH3OOH_HV_IRR: CH3OOH->[j]CH2O+HO2+OH
CH2O_HV_IRR: CH2O->[j]HO2+HO2+CO
CH2O_HV_a_IRR: CH2O->[j]CO+H2
H2O2_HV_IRR: H2O2->[j]OH+OH
CH3CHO_HV_IRR: CH3CHO->[j]CH3O2+CO+HO2
POOH_HV_IRR: POOH->[j]CH3CHO+CH2O+HO2+OH
CH3COOOH_HV_IRR: CH3COOOH->[j]CH3O2+OH
PAN_HV_IRR: PAN->[j]0.6*CH3CO3+0.6*NO2+0.4*CH3O2+0.4*NO3
MPAN_HV_IRR: MPAN->[j]MCO3+NO2
.....................
You just need to add the comment, species_list and species_group_list to the yaml file.
For more mechanism yaml files, please check the my forked branch.
Version control
| Version | Action | Time |
|---|---|---|
| 1.0 | Init | 2020-12-02 |
Say something
Thank you
Your comment has been submitted and will be published once it has been approved.
OOPS!
Your comment has not been submitted. Please go back and try again. Thank You!
If this error persists, please open an issue by clicking here.